In recent years, research on the qualitative improvement of endomicroscopy using Artificial Intelligence has seen our group contribute both with new methodologies and systematic studies. Below are all main published works, with a clear note on what each contribution introduces.
Motivations & Objectives
- Raise the diagnostic quality of endomicroscopy images using super-resolution techniques, enabling reliable clinical applications even in the absence of high-resolution data.
- Compare, standardize, and bring to the field advanced deep learning techniques for SR and spatiotemporal reconstruction.
Methods and Results
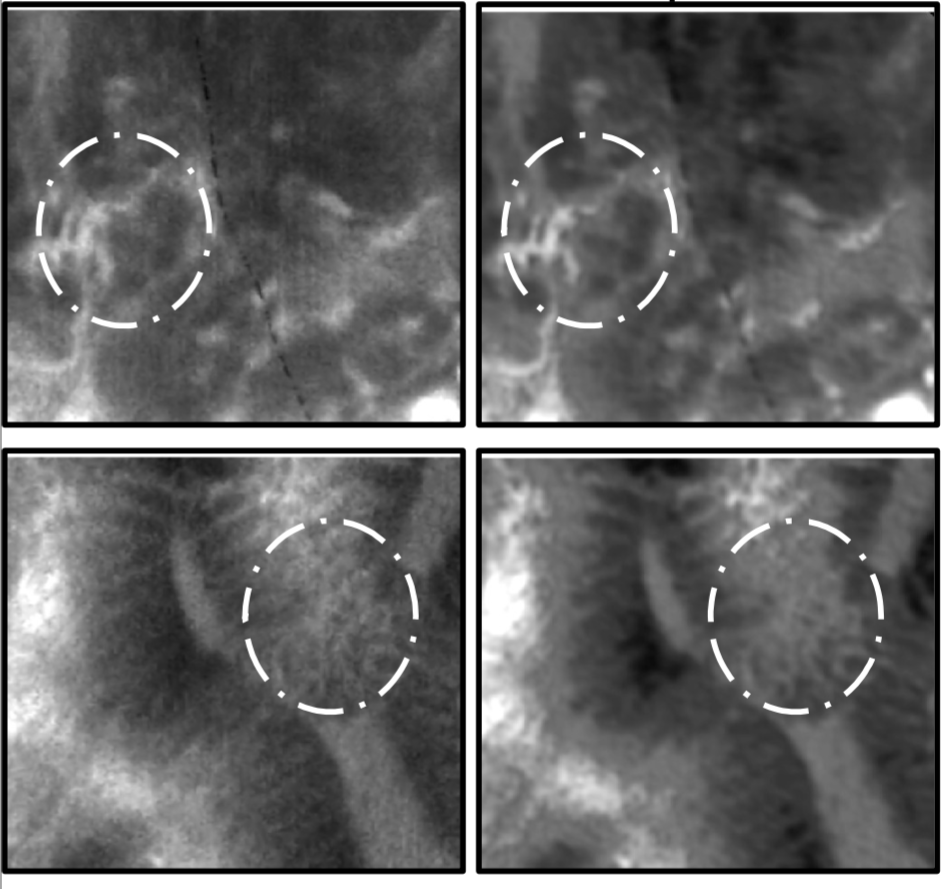
- Adversarial training with cycle consistency for unsupervised super-resolution in endomicroscopy
Medical Image Analysis, 2019The first unsupervised framework based on GAN and physically-inspired cycle consistency: allows SR learning even in the absence of perfectly aligned LR/HR pairs, transferring HR quality between different domains. Validated on 238 video sequences from 143 patients, with quantitative metrics and clinical MOS study.
- Effective deep learning training for single-image super-resolution in endomicroscopy exploiting video-registration-based reconstruction
International Journal of Computer Assisted Radiology and Surgery, 2018 –
PMC Open AccessProposes an effective strategy for synthetic data generation via video-mosaicking to train DNNs for SR even when real HR images are lacking. Analyzes three state-of-the-art techniques on over 8,800 images and confirms (also with MOS) the qualitative improvement.
- Learning from irregularly sampled data for endomicroscopy super-resolution: a comparative study of sparse and dense approaches
International Journal of Computer Assisted Radiology and Surgery, 2020Comparative study of dense vs sparse CNN for super-resolution from irregular inputs (typical of pCLE); also introduces Nadaraya-Watson kernel layers into the deep framework and generates synthetic data for robust benchmarking.
Code Repository
- Currently, dedicated open-source code for endomicroscopic super-resolution is not available in public repositories.
For requests or collaborations, contact the authors of the cited works.
Team & Authors
- Daniele Ravì (PI, deep learning methodologies, GAN/SR framework design, real data validation)
- Agnieszka B. Szczotka (idea and development of sparse/dense CNN, dataset, UCL validation)
- Stephen P. Pereira, Tom Vercauteren, D.I. Shakir, M.J. Clarkson (KCL, UCL, clinical collaboration and supervision)
Collaborations UCL–King’s College London, clinical validation on patients, benchmarking on own and open databases.