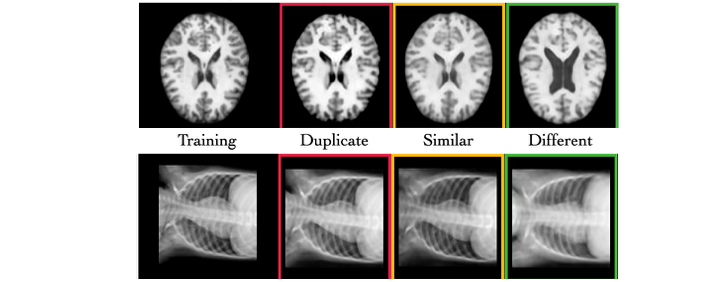
The diffusion of deep generative models has led to the creation of synthetic data for medical imaging, which is essential for the research and development of new diagnostic techniques. However, these models seriously risk “memorizing” sensitive patient data, generating images that are too similar to the training data and thus exposing confidential information. The DeepSSIM project was created to address this critical issue, proposing a new automatic and self-supervised metric to quantify how much a generative model memorizes original data during synthesis.
Motivations & Objectives:
- Protect patient privacy in the age of medical AI.
- Provide a practical tool to evaluate the risk of memorization in generative models used in healthcare.
- Promote a culture of transparency and reproducibility in research on sensitive data.
Methods:
- DeepSSIM projects images into a neural embedding space for learning, optimizing cosine similarity to reflect the true SSIM (Structural Similarity Index) between medical images.
- The algorithm uses augmentations that preserve anatomical structure, enabling similarity estimation without requiring perfect spatial alignment.
- Validated on synthetic MRI images generated by a Latent Diffusion Model trained under high memorization risk conditions.
Impact & Results:
- DeepSSIM outperforms all previous metrics with a +52.03% average F1 score increase in memorization detection.
- Ensures efficient and scalable analysis even on large datasets, speeding up calculations tens of times compared to traditional SSIM.
- Open-source code available for the community, fostering transparency and future applications.
Related Scientific Articles
- A Novel Metric for Detecting Memorization in Generative Models for Brain MRI Synthesis
(arXiv preprint arXiv:2509.16582, 2025) –
Google Scholar
Code Repository
- Official repository: brAIn-science/DeepSSIM
Team & Authors
- Antonio Scardace (First author, methodology development)
- Lemuel Puglisi (Co-author, implementation)
- Daniele Ravì (Scientific supervision)
- With Francesco Guarnera, Sebastiano Battiato.